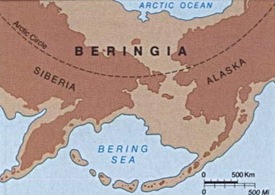
 If you’re interested in DNA, Native American History, or genetic genealogy, then you’re undoubtedly heard of a new paper from PLoS ONE called “The Phylogeny of the Four Pan-American mtDNA Haplogroups: Implications for Evolutionary and Disease Studies.” The authors, from all around the world (including Ugo A. Perego from SMGF and Antonio Torroni from Italy) analyze over 100 complete Native America mtDNA genomes. From the abstract:
If you’re interested in DNA, Native American History, or genetic genealogy, then you’re undoubtedly heard of a new paper from PLoS ONE called “The Phylogeny of the Four Pan-American mtDNA Haplogroups: Implications for Evolutionary and Disease Studies.” The authors, from all around the world (including Ugo A. Perego from SMGF and Antonio Torroni from Italy) analyze over 100 complete Native America mtDNA genomes. From the abstract:
“In this study, a comprehensive overview of all available complete mitochondrial DNA (mtDNA) genomes of the four pan-American haplogroups A2, B2, C1, and D1 is provided by revising the information scattered throughout GenBank and the literature, and adding 14 novel mtDNA sequences. The phylogenies of haplogroups A2, B2, C1, and D1 reveal a large number of sub-haplogroups but suggest that the ancestral Beringian population(s) contributed only six (successful) founder haplotypes to these haplogroups.”
 On the heels of my recent post discussing all the interesting information that’s recently entered the blogosphere about genetic genealogy and DNA studies, here are a few more:
On the heels of my recent post discussing all the interesting information that’s recently entered the blogosphere about genetic genealogy and DNA studies, here are a few more: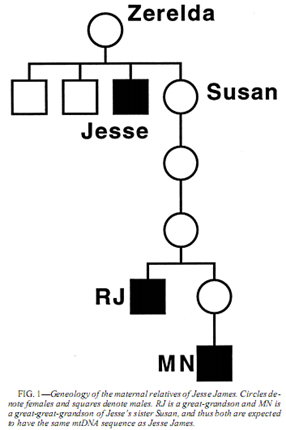 Jesse Woodson James, born September 5, 1847 and died April 3, 1882, was an infamous American outlaw. Despite strong evidence that James was killed on April 3, 1882, some theorized that his death was staged and that he in fact survived to father additional children.
Jesse Woodson James, born September 5, 1847 and died April 3, 1882, was an infamous American outlaw. Despite strong evidence that James was killed on April 3, 1882, some theorized that his death was staged and that he in fact survived to father additional children. As I was reading through the GENEALOGY-DNA list from Rootsweb this morning, I came across a great
As I was reading through the GENEALOGY-DNA list from Rootsweb this morning, I came across a great  A report published in the New England Journal of Medicine entitled “
A report published in the New England Journal of Medicine entitled “ As of the end of November, the Personal Genome Project has a newly-designed and user-friendly
As of the end of November, the Personal Genome Project has a newly-designed and user-friendly