 Short Summary: Before the end of the year, AncestryDNA plans to update our match lists using a new algorithm that reduces the number of false positive matches. For the first time, matching DNA segments will be characterized as IBS (i.e., a false positive) based on something other than simply segment length.
Short Summary: Before the end of the year, AncestryDNA plans to update our match lists using a new algorithm that reduces the number of false positive matches. For the first time, matching DNA segments will be characterized as IBS (i.e., a false positive) based on something other than simply segment length.
AncestryDNA Day
Last Monday, October 6th, I and six other members of the genetic genealogy community attended a ‘Bloggers Day’ hosted by AncestryDNA at the San Francisco headquarters of Ancestry.com. Two other members of the group have already written about the event:
- When less is more – The Legal Genealogist by Judy Russell
- DNA Day With Ancestry – DNAeXplained by Roberta Estes
While at ‘Bloggers Day’ we discussed many issues including the Y-DNA and mtDNA databases originally scheduled for destruction, upcoming changes to AncestryDNA’s matching algorithm (much more below), and other upcoming changes to the AncestryDNA about which you will hopefully soon hear much more.
I would like to thank Ancestry.com for reaching out to the genetic genealogy community, for organizing this event, and for allowing us to share information with our readers. I think this is the start of a very positive line of communication and collaboration, and I look forward to the future.
Reducing False Positives
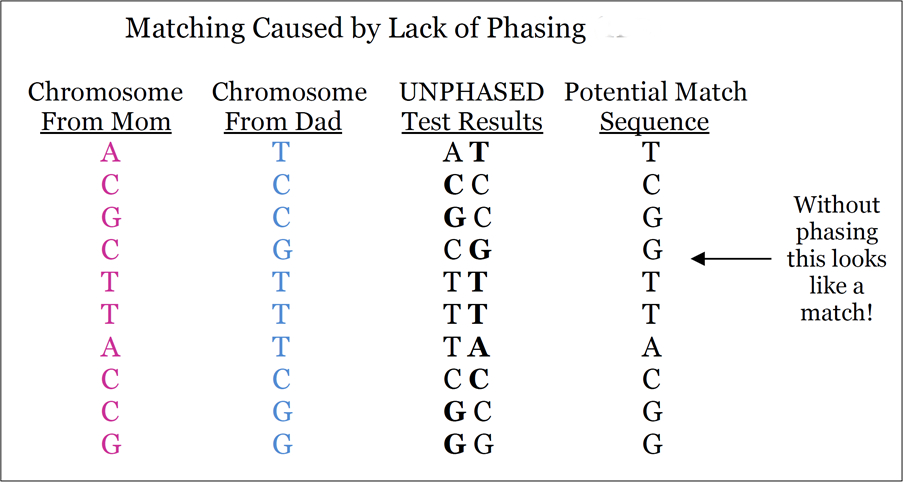
Currently, identifying genetic cousins is far from a perfect process. Many of our “matches” are actually false positives, meaning that the two or more individuals share a segment of DNA in common for a reason other than via recent inheritance from a common ancestor. For example, they could share a segment of DNA as a result of being a member of a particular population rather than due to a recent common ancestor, or that could actually not share the segment of DNA (for example, matching that results from a lack of phasing or errors in phasing, among other reasons).
With this in mind, there are currently several ways to reduce the number of false positives:
Phasing:
Many false positives result from the lack of phasing (see the figure below). Although using DNA from family members (such as a “trio” or “triad” of a child and two parents) is the best way to phase a genome, researchers have long been phasing genomes using large datasets and algorithms (see the software list HERE and an aging review article HERE). While not perfect, this method of phasing reduces the number of false positives that result from unphased data. Unfortunately, phasing individuals using data from family members is both computationally expensive and not very scalable.
AncestryDNA has been working on improvements to their phasing algorithm, which will be a component of the update later this year. The new algorithm reduces the error rate by as much as 66%, and I expect to see further improvements as the database grows.
Filtering by Segment Size:
By far the most common method for reducing false positives is to set a minimum threshold for the length of a matching segment. The longer a segment shared by two individuals, the more likely it is that the segment was mutually inherited from a common ancestor (i.e, that the segment is Identity-by-Descent or IBD) rather than shared for some other reason (i.e., that the segment is Identity-by-State or IBS).
23andMe, for example, sets a minimum threshold for a first segment of 7 centiMorgans (“cMs”) and 700 SNPs. AncestryDNA has a threshold of 5 cMs for the first shared segment. Family Tree DNA sets a threshold of 7.7 cMs for the first segment. In other words, two individuals must share at least one segment of DNA that satisfies the minimum threshold set by the testing company in order for the individuals to be considered genetic matches.
Unfortunately, there is insufficient data regarding the relationship between segment size and IBS versus IBD. Earlier this year, 23andMe published research entitled “Reducing pervasive false positive identical-by-descent segments detected by large-scale pedigree analysis” that describes an open-source algorithm called HaploScore. HaploScore provides a metric by which to rank the likelihood that a stretch of DNA is inherited IBD between two individuals or not. In the paper, however, the researchers determined a false positive rate of over 67 percent for short (2 to 4 centiMorgans) segments of DNA. In other words, for every 4 cM segment shared by two individuals, there is a 67% chance that the segment is IBS rather than inherited from a common ancestor. Based on Figure 2(b) of the paper, it appears that the false positive rate for 5 cM segments is approximately 30%.
The “Autosomal DNA statistics” page at the ISOGG wiki provides some estimates of IBS v. IBD based on length, although neither the underlying data nor the method used to obtain this information were shared. This data suggests that a shared segment of DNA having a length of 5 cM has a 95% chance of being IBS. In other words, for every 100 five cM segments, only five might be real. Again, however, the method and dataset have not been peer-reviewed (and the results do not align well with the paper discussed in the previous paragraph).
Accordingly, there is certainly a relationship between length and IBS versus IBD, and while the companies set a threshold in an attempt to reduce false positives, length alone is not sufficient to weed out all false positives. Indeed, some have reported from their analysis of their own triad that even rather long segments – on the order of 10 cMs or longer – are false positives.
AncestryDNA’s New Matching Method – Incorporating Allele Frequency
NOTE: the following information is based on my understanding of the new matching algorithm. There will be a white paper that is released later this year along with the update, so please read that white paper for the official details and explanation. Further, please note that these figures below are research data and won’t necessarily be a part of the user experience once the matching algorithm is implemented.
In order to reduce false positives that fall above the matching threshold, AncestryDNA has leveraged the immense amount of information about allele frequency in their database in order to modify their matching algorithm.
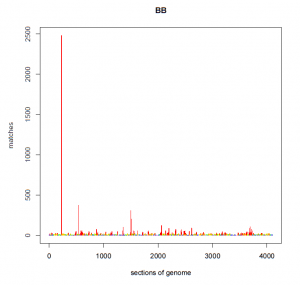
For example, below is a graph of the number of genetic matches (Y-axis) that share a particular segment of DNA in my genome (X-axis). What you will immediately notice is that at one location of my genome, I share a segment of DNA with 2,500 other people. At two other locations, I share DNA with 300 to 400 people, which is significantly higher than every other segment of my DNA. Unfortunately I do not know where these segments are found within my genome, but I might be able to guess using information from Family Tree DNA and/or GEDmatch (where many of us have noticed these “pileups” previously).
BEFORE:
Statistically, the likelihood of sharing the exact same segment of DNA with 2,500 other people in the database (0.5% of the 500,000 people in the database) is almost zero. Accordingly, this segment of DNA is not shared among all of these matches because of mutually shared recent ancestry, but is shared due to one or more other factors.
Thus, AncestryDNA is identifying the frequency of certain alleles in the database and using that information to reduce false positives. I certainly don’t want to see those 2,500 matches (at least not if the peak is our only shared DNA), or either of those two groups of 400 matches. I currently match about 9,700 people at AncestryDNA – eliminating the 3,300 false positives in just those three peaks will be a good start.
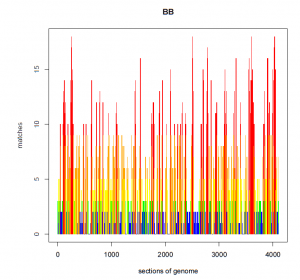
After applying the allele frequency algorithm to my data, my graph looks like the one below, where at every position along my genome I have fewer than 20 matches.
AFTER:
Thus, size and allele frequency can be combined to eliminate false positives. A shared segment of DNA, if it appears at a significantly high frequency in the database, is much more likely to be IBS than IBD, even if it is a large segment. Similarly, a shared segment of DNA that is less frequent in the database is much more likely to be IBD than IBS, even if it is a very small segment. Accordingly, small segments of DNA can be preserved in matching but because of the new algorithm will tend to be IBD rather than IBS.
For the first time, matching will be based on a factor other than segment length.
Important issues
In my mind, there are several important issues that this algorithm raises, which hopefully will be addressed by the white paper that accompanies the update. For example:
- Endogamous populations: I am not certain how endogamy will be treated by this algorithm, although I will expect it to be covered completely in the white paper. We were told that this will benefit endogamous populations, but again I am not sure exactly what the impact will be, particularly if the ethnicity estimate is not a factor in matching. It is easy to envision scenarios on both sides of the fence.
- Confirmation: Unfortunately, the changes implemented by AncestryDNA will be very difficult to confirm using third-party tools such as GEDmatch, because these tools will not have access to the underlying allele frequency information available to AncestryDNA. Although GEDmatch or other third-party tools will be able to analyze allele frequency (indeed, many genetic genealogists have already identified problematic areas of their genome that are shared by too many other people), these databases are smaller and won’t have nearly as much information about allele frequency. What this means, unfortunately, is that people will find segments at GEDmatch that they share with other AncestryDNA test-takers who are not reported to be their genetic cousin, and may overlook the possibility that the shared segment was intentionally ignored due to its frequency per the above analysis. In other words, people using GEDmatch WILL find shared segments and matches that are [intentionally] not reported by AncestryDNA. These matches and/or segments are intentionally not reported by AncestryDNA because they are false positives. It will be important to consider the possibility that these unreported segments are over-represented (and therefore false positive) alleles.
Preparation
The AncestryDNA team expressed interest in making a download of original matches available to users even after the algorithm update. In other words, the download would include a list of everyone that the test-taker ever matched (including the thousands of false positives). Although I believe that using your list would be a colossal waste of time because anyone found in the download and not in your revised match list will be a false positive, I’m not sure that I would recommend against downloading it, at least for now.
Note that there is no guarantee that such a download will be available, it was simply discussed as a possibility, and all the attendees strongly supported the idea.
Conclusions
There is no question that this algorithm will have a significant impact on our match lists. I expect the implementation of this algorithm to reduce matches by as much as 60 to 90%. Accordingly, I expect my matches to go from 9,700 to closer to 1,000 (and possibly even less). I feel very confident that the missing matches will be false positives, although of course I will need to read the white paper in order to further educate myself.
AncestryDNA expects to implement the algorithm and update match lists before the end of 2014.
Before I finish, let me very clear: I want the underlying segment data from AncestryDNA. Although the vast majority of AncestryDNA users would never utilize that segment data, I believe that providing it would lend additional credibility to the product as well as foster greater trust and collaboration between the company and the professional and advanced users. It can do nothing but good. Although there are valid privacy concerns involved (for example if I share a segment of DNA with you and I know that DNA contains a certain disease-causing mutation, I know you likely have the same disease-causing mutation), I am not aware of anyone ever intentionally using segment data to determine a match’s health status. Further, segment data could be an opt-in system or could be shared only after the test-taker reviews and accepts an updated informed consent document. There are a number of ways around the privacy issues.
Even absent the underlying segment data, I believe that the new algorithm is a very good development in the ongoing War Against IBS.
Full Disclosure: AncestryDNA arranged for travel, hotel, and other expenses for this summit. To help cover the costs associated with blogging and my genetic genealogy educational efforts, I offer affiliate links for all three testing companies on the blog.
.
“AncestryDNA has a threshold of 5 cMs for the first shared segment.”
I believe I read that AncestryDNA actually has set the threshold to 5 Mb (not cM) for those that tested at an earlier date and newer matches are calculated in cM’s. Was there any word if the entire database will be switched over to matching by cM’s and if so when might this occur?
Dean – I don’t believe there was any mention of this at the meeting. Hopefully they will address this issue in the forthcoming white paper that accompanies the update, as I imagine it will have to be considered.
Blaine: Thanks for the write-up
I will quibble with this little section: ” Statistically, the likelihood of sharing the exact same segment of DNA with 2,500 other people in the database (0.5% of the 500,000 people in the database) is almost zero. ”
Rather, it appears the likelihood is actually 1, not 0. The fact is, AncestryDNA has *measured* that you are sharing a tiny section of a chromosome with 2500 people. So the question is *why*, not *if* .
These traffic jams on portions of chromosomes has, as you noted, been seen by many people. Using data from FTDNA and gedmatch.com I’ve seen that my mother has pile-up on the HLA portion of chr6. This is almost certainly from some deep European common ancestry.
Interesting, perhaps if I added “in a genealogically relevant timeframe” to the sentence it would help, although I’m also hypothesizing that a substantial fraction of those matches are completely IBS caused by phasing errors, etc. (i.e., not old IBD).
Currently, 60% – 90% of AncestryDNA’s matches are false. Based on what people at AncestryDNA has reported, I think this is a reasonable estimate:
http://33.media.tumblr.com/f79b70e4d6130e287c3e6aeaaa41d6c6/tumblr_nd4ils45qD1txt4yho1_1280.png
If the slide is any indication, match lists will be dropping 60 -90% after the purge.
I’m glad ancestry is cleaning things up, but a 60 – 90% false positive rate raises questions about how they managed to do so poorly in the first place.
I hope AncestryDNA knows what they’re doing this time.
Jason – it was an intentional choice to be more inclusive rather than more exclusive, and I think that was the right choice. Of course the goal is for there to be neither false positives nor false negatives, but we’re not there yet. So better to include false matches than to miss real matches.
And keep in mind these false positives caused by pileups will exist across ALL the companies and all the databases, so technically everyone is doing “so poorly.” So far only AncestryDNA is using this approach to weed them out.
Are you saying they are going to take away some of the matches presently in our accounts? If so, okay by me, since I have been through ALL of my many thousands of trees, and keep current with each day’s new additions of approx. 25. I have a notebook filled with my matches. I have found 85 dna matches who also have Stephen Hopkins b1581 in their trees. But, those of us who descend from Colonial America, would not many of us be descended from him?? Of course, I know without segment matching, that is not a match. But, I would opine most of the other matches do not know that, because AncestryDna has not bothered to educate them.
I am not sure what I am doing, or where I am going, but………I am on my way. LOL
Caith,
That’s right, you’ll see a serious reduction in your matches, which most of us welcome. Yes, you’re right, of course. Some have hypothesized, for example, that Mayflower immigrants John and Priscilla Alden may have a million living descendants or more, but only a tiny, tiny fraction will have any of their DNA.
I don’t know why Ancestry doesn’t offer manual phasing, like GEDMatch. We could phase our own kits if we have parent and child tests, and then those phased kits could be used in cousin matching instead of the unphased kits. And if the Ancestry software could indicate which side of the family a given cousin match is coming from, it would be very helpful. Not every person on Ancestry is also on GEDMatch, so finding out who that “probable 4th cousin match” is related to in your pedigree can be quite a challenge.
Well, even after your DNA is phased by the AncestryDNA algorithm, they don’t know which chromosome is your mother’s and which chromosome is your father’s. The best they could do is say that certain matches are on #1 of chromosome 21, and certain matches are on #2 of chromosome 21. That would be immensely helpful, of course, but it would be up to us to figure out whether #1 is mom or dad.
I am very excited to hear about the purge. I have over 18,000 matches at AncestryDNA and it is overwhelming! I do use their surname search and shared hints sorting to make it more manageable, but getting rid of 6,000+ matches would be good.
It would be very nice if Ancestry gave us access to the segment data. FTDNA does it. And though Ancestry seems to be overly worried about privacy, as you mention, there are ways to opt in our out. I just hope they make opt in the default if they come around.
But I’m still concerned about 5cM as the lower threshold. I think that’s too low and is responsible for a great many of my false positives. If they are not willing to let us download the shared segment data, I hope they at least could give us some controls to filter by setting our own thresholds. I’d love to be able to set it at 10cMs so I can work on the higher probable matches first. Once you get into the 5th-8th cousins section it’s a crapshoot.
Is it possible for others to get their segment info from AncestryDNA like you did?
Thanks for your summary.
Jackson – it isn’t, unfortunately. And even if you could, it isn’t very useful since it doesn’t include the exact locations of the pile-ups. For example, although I know I had big pile-ups in my genome, I have no idea where they were located.
I had my test done at Ancestry and the composition was NOT as reported by my family. Specifically I AM SURE that my great great grandmother on my mother’s side was of Irish ancestry and equally AS SURE that my father’s father is a Cherokee! Well when I got my results from ancestry I was reported as less than 1 percent native american, and none of my european ancestry was Irish!
Not convinced of Ancestry.com accuracy I did the free upload and free 1st 20 matches from Family Tree DNA and VIOLA one of my first two matches is of total Irish ancestry, and a few down show North Carolina/Virginia area which is traditionally Cherokee Nation. Whilst DNA is DNA ancestry.com racial break down information was ALL WRONG for me!
You can transfer your raw data to FTDNA using this link for free https://www.familytreedna.com/autosomalTransfer?atdna=mGuGbpp1PgJxZC6kBS9UEg%3d%3d
Please do tell if the ancestry information is more accurate with FTDNA that with Ancestry.com