 Today, The Genographic Project officially announced the launch of their new Geno 2.0 project, a significant update to the type and quantity of genetic information that will be collected and analyzed by The Genographic Project. The new project will use an entirely new SNP chip (the GenoChip) designed specifically for Geno 2.0 in order to provide the world’s most detailed information about Y-DNA and mtDNA haplogroups (using SNP information) as well as detailed biogeographical estimates and ancient population (Denisovan and Neanderthal) estimates.
Today, The Genographic Project officially announced the launch of their new Geno 2.0 project, a significant update to the type and quantity of genetic information that will be collected and analyzed by The Genographic Project. The new project will use an entirely new SNP chip (the GenoChip) designed specifically for Geno 2.0 in order to provide the world’s most detailed information about Y-DNA and mtDNA haplogroups (using SNP information) as well as detailed biogeographical estimates and ancient population (Denisovan and Neanderthal) estimates.
As of today you can pre-order a Geno 2.0 kit, which is expected to ship no later than October 30th (although you can probably expect it earlier than that).
Once again Family Tree DNA will perform all the testing, and The Genographic Project has worked very closely with FTDNA to design, troubleshoot, and use the GenoChip. FTDNA will perform both the Family Finder and the Geno 2.0 test.
Overview of Geno 2.0
The new Geno 2.0 SNP chip contains roughly the following SNPs:
- ~3,200 mtDNA SNPs
- ~12,000 Y-DNA SNPs
- ~130,000 autosomal and X-chromosomal AIMs (including ~30,000 SNPs from candidate regions of interbreeding between extinct hominins (Denisovan and Neanderthal) and modern humans)
The AIMs (Ancestry Informative Markers) were derived from roughly 450 populations around the globe, including many that are unique to the Genographic project and many that have never been previously searched for AIMs. The SNPs in regions believed to represent Denisovan and Neanderthal interbreeding will be used to detect and study DNA flow between humans and these extinct populations.
Overview of Results
So what do you get when you order a Geno 2.0 test? Via the new user interface (some of which you can see below), you will receive the following:
- A deep-clade Y-DNA haplogroup assignment;
- A deep-clade mtDNA haplogroup assignment;
- Information about the history and migration of mtDNA and Y-DNA haplogroups;
- A biogeographical (ethnicity) estimate; and
- An ancient population (Denisovan and Neanderthal) estimate.
What Geno 2.0 does NOT do:
- Geno 2.0 does not reveal medically-relevant information (but note that medical/health/trait information can sometimes be revealed unintentionally as new health associations are discovered, for example). NG went to great lengths to prevent medical/health/trait information from being detected by the Geno 2.0 chip. For example, the team selected only non-coding SNPs with no known functional association, and filtered all selected SNPs against a 1.5 million SNP database (which they constructed from numerous sources) containing all SNPs known or believed to be associated with disease or health. The team also removed all SNPs with a high association with medically-relevant SNPs (which you might be familiar with b/c of Dr. Watson and his APOE status).
- Identify genetic cousins with autosomal DNA. The Geno 2.0 product is not intended to identify close genetic relatives based on autosomal DNA, and thus does not have that functionality. I’m guessing that it will be possible for third-party sites to glean some information about relatedness from the data, however. Note that relatedness through the Y-DNA or mtDNA based on haplogroup information is a part of the functionality of Geno 2.0, as can be seen in the screenshots below.
FTDNA (Deep Clade Testing; Integration of Genographic Results)
One interesting aspect of the new Geno 2.0 chip is that it will completely replace the deep-clade analysis performed at FTDNA. With this one test, all SNPs currently analyzed by FTDNA in all of its different deep-clade analyses are analyzed in their entirety.
Another great benefit of the Geno 2.0 test is that Genographic will allow the test-taker to upload/transfer their results back into FTDNA, and neither Genographic or FTDNA will charge a fee for this transfer. This means that Project Administrators will be able to work within the FTDNA system to analyze results of their project members rather than having to rely on collecting data from project members outside the system (thereby potentially increasing participation and results).
Validation of the GenoChip
The designed GenoChip has undergone significant validation (including the use of about 400 known Y-DNA and mtDNA samples, and as many as 650 samples from various populations around the world). Following this validation process (which will continue for at least the foreseeable future), the validated GenoChip SNPs are as follows:
- ~12,000 Y-DNA SNPs
- ~3,200 mtDNA SNPs
- ~130,000 autosomal and X-chromosomal SNPs which include the following SNPs:
- 23,692 Neanderthal
- 1,357 Denisovan
- 12,027 Aboriginal
- 10,159 Eskimo Saqqaq
- 998 Chimpanzee
Downloadable Raw Data: Treasure for Third-Party Analysis and Apps
The test-taker’s raw data will be available for download by the test-taker. This has happily become the norm for most genetic genealogy companies, and NG will follow suit. It’s not clear at this point whether that will be an immediate functionality (although I’m guessing it will be), or whether it will be in the near future.
Interestingly, the ability to download raw data opens the door for third-party analysis. For example, I data from the Geno 2.o chip will lead to significant new mtDNA and Y-DNA discoveries (using the user-fueled Y-Chromosome Genome Comparison project, for example).
Consent
During a presentation put on by Spencer Wells and FTDNA a few weeks ago describing the new Geno 2.0 project, I and several other DNA bloggers were able to ask questions about the new chip and the project. I and a few others asked questions about consent, which is of course an important aspect of any research project involving human samples.
Specifically, I asked whether all test-takers are automatically participants in the research aspect of the new Geno 2.0 project. Dr. Wells responded that test-takers must opt into research; they are not automatically research participants. Accordingly, people who are interested in the new test but have concerns about participating in research can do so.
The Geno 2.o Terms and Conditions are here.
Miscellaneous
A few miscellaneous points:
- As of the recent presentation, Genographic was not yet certain if they will be storing DNA samples after they are tested. They are considering doing so, but of course there are significant costs associated with long-term storage of tens of thousands of DNA samples.
- Testing will take approximately 4-6 weeks once the system is in full swing (but I’m guessing there might be some delays in the beginning with an initial influx of orders).
- You will eventually be able to order the Geno 2.0 test directly through the FTDNA website.
- Although not completely reflected in the screenshots below, the new project allows for much greater participant involvement and interaction. For example, test-takers are encouraged to share their stories after receiving their results.
For More Information:
Several other bloggers will be writing about today’s launch, and I will update this post to include links to those reviews. Also, since I will be taking the Geno 2.0 test in the near future, stay tuned for my review and results.
- National Geographic and Family Tree DNA Announce Geno 2.0 – at Your Genetic Genealogist (CeCe Moore)
- Geno 2.0 launches! – at The Legal Genealogist (Judy G. Russell)
- The Genographic Project: onto the autosome! – at Gene Expression (Razib Khan)
- National Geographic – Geno 2.0 Announcement – The Human Story – at DNAeXplained (Roberta Estes)
- National Geographic Announces New DNA Test – DNA – Genealem’s Genetic Genealogy (Emily Aulicino)
Screenshots
Below are a series of screenshots from the new Geno 2.0 project and test results, provided by National Genographic:
The complete kit (collection is by cheek swab):
Introduction Page:
Overview Page:
An mtDNA haplogroup and heat map (showing modern-day locations and frequencies of the haplogroup):
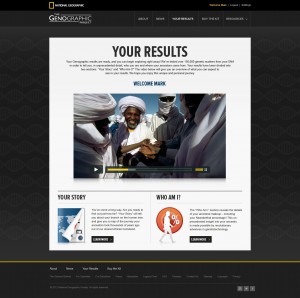
The “Your Story” main page:
An example of more information at the “Your Story” page:
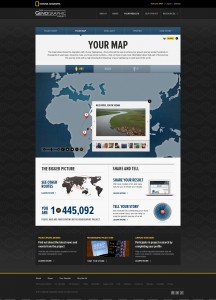
The “Your Map” page (showing an mtDNA map):
More of the “Your Map” page (showing an mtDNA map):
NOTE: much of this information is based on preliminary information about the GenoChip and Geno 2.0 project. Accordingly, the information is subject to change. Check The National Genographic website for the latest information.






Cece has attributed this quote to you. I don’t know when or where it is from.
I personally see many uses, including ethnicity prediction (which interests both genealogists and non-genealogists); deep clade haplogroup analysis (something I’ve been meaning to do for years but never got around to); SNP discovery to refine Y-DNA and mtDNA trees, etc. I think there may also be numerous third-party uses, some of which we haven’t thought of yet. It’s not useful for identifying autosomal cousins, but of course there’s much more to genetic genealogy than identifying cousins.
Perhaps you could elaborate on this part of your comment?
“….but of course there’s much more to genetic genealogy than identifying cousins.”
Identifying cousins using autosomal DNA testing has only been around for about 3 or 4 years since 23andMe and FTDNA launched their Relative Finder and Family Finder products. Before that some people used Y-DNA and mtDNA to find or disprove relationships, but many people undergo genetic genealogy simply to learn about their ancient ancestry: their Y-DNA haplogroup and origins, their mtDNA haplogroup and origins, and their biogeographical (ethnicity) estimate. The Geno 2.0 test will do those three things together better than any current test.
Thus, while identifying genetic cousins is an important goal of many genetic genealogists, a large percentage of test-takers have other interests.
Hi Blaine,
As you probably know, the quote was a comment you made on my blog.
Thanks for the link and nice post, as usual!
Blaine,
Thank you for your reply. Not that anyone wouldn’t be curious about such things,but I didn’t think that haplogroup and biogeographical (ethnicity) estimates were possible – yet. Given that DNA is remixed over generations by multiple populations, at least in current Western populations. Are you saying that both the time and and biogeographical periods will actually be evident in this test? Aren’t the time spans given currently only in 1000 year increments? If so, how could an ethnicity or near geographical position be evident in anything less than generalities?
I have had my 37 dna test with familytreedna why do i need another another kit? and will the results be made known as far as possible in lay mans language?
also will it clarify what haplogroup ones belongs to?
These test while they are interesting are geared to those people that are over the top obsessed with seeing their ancestry broken down into percentages. Considering most of these folks have already tested with a plethora DNA testing
companies as if by know they don’t know what or who they are? As if people need to know where they came from 10,000 years BC.
However, get ready for the hordes of the smaller portion of humanity to shell out another $300 dollars who are by now addicted. Like rolling a dice……What ancestry will come out this time. Will Cherokee grandma’s
DNA come out this time…..Shake it up baby…..let her rolllll!!!!
Awesome Look. Do they test haplogroups as well with the new chip? If so, how many SNP/STR haplogroup markers do they scan? I’ll definitely try it out later.
“Another great benefit of the Geno 2.0 test is that Genographic will allow the test-taker to upload/transfer their results back into FTDNA, and neither Genographic or FTDNA will charge a fee for this transfer. This means that Project Administrators will be able to work within the FTDNA system to analyze results of their project members rather than having to rely on collecting data from project members outside the system (thereby potentially increasing participation and results).”
This is not true, ftdna would not allow me to transfer my mtdna results because of the chip:
Hello,
Thank you for contacting the Genographic Project. The only information that would have been transferred would be found under the mtDNA>Haplogroup Origins section of your FTDNA profile. Since you do not have any mtDNA matches with our database, you do not have any information at this time. The data-set for mtDNA is not compatible with FTDNA. The mtDNA portion of the Geno 2.0 test is run on a chip, whereas FTDNA must individually sequence strands of your mitochondrial DNA. It is possible this information will be transferable in the future.
Have a nice Day
Regards,
Clint
Family Tree DNA
http://www.familytreedna.com
History Unearthed Daily
I was excited by the Genographic 2.0 offer. I ordered my kit in early December and mailed back my swabs for testing before Christmas. I eagerly awaited the 6-8 weeks to get my results…and waited…and waited…and waited. It has now been 5 MONTHS and the online results page says my analysis is only 60% completed. I contacted National Geographic for a partial refund because of the exceedingly long delay, but they refuse to issue one. This is a new and apparently popular test. I would urge people to seek out another DNA testing company than National Geographic, their Genographic Project, and their contractor Family Tree DNA unless you are willing to wait half a year or more for your results.
Shame there’s not an option to use your FamilyTreeDNA test results in this project. A friend of mine spent over $500 for the Comprehensive Genome test and would love to be able to use the results for this project instead of having to spend another $200 if he wants to join this one.
I find the science behind the Geno 2.0 Project a little flawed. Believing that we evolved from a fish to a reptile to a chimp to a Neandrathal man to a human is alot of big jumps with NO REAL PROOF that this is what happened. The migratory routes extending out of Africa to the Mideast and then on to Asia and Europe are seriously in question. Where is the real science that this is exactly what happened? I tend to believe that it all started in Mesopatamia and then in branched out from there. Prove me wrong! I don’t doubt what DNA can do in genealogy to some extent however these huge leaps in evolution are just theories and not real science — thus the Theory of Evolution. I believe the oldest history book in the world when it indicates that races and languages evolved after the Tower of Babel. The scientist(s) behind the National Geographic Genographic Project should give their complete and connected science behind their logic or otherwise indicate in the results that their migratory routes and Neonadrathal/chimp genes are part of theories too that cannot be proved by real science.
John, I fully agree, my friend! I consider the Creator G-d the ultimate expert in the development of the mankind He made in His likeness! G-d bless you!